Overview
Visualise the results of F test to compare two variances, Student’s t-test, test of equal or given proportions, Pearson’s chi-squared test for count data and test for association/correlation between paired samples.
Installation
# CRAN installation: install.packages("gginference") # Or the development version from GitHub: # install.packages("devtools") devtools::install_github("okgreece/gginference")
Usage
One sample
One sample t-test with normal population and σ2 unknown
The rejection regions for one sample t-test with normal population and σ2 unknown are calculated using ggttest function. The following table shows the rejection regions which is calculated with gginference depending the specified parameters in t.test.
| H0 | H1 | Rejection Region of gginference |
Parameters of t.test
|
|---|---|---|---|
| μ = μ0 | μ < μ0 | R = {z < − za} |
|
| μ > μ0 | R = {z > za} |
|
|
| μ ≠ μ0 | R = {|z| > za/2} |
|
where

One sample t-test with normal population and n < 30 and σ2 unknown
ggttest is also used to calculate rejection region for one sample t-test with normal population and n < 30 and σ2 unknown.
| H0 | H1 | Rejection Region of gginference |
Parameters of t.test
|
|---|---|---|---|
| μ = μ0 | μ < μ0 | R = {t < − tn − 1, a} |
|
| μ > μ0 | R = {t > tn − 1, a} |
|
|
| μ ≠ μ0 | R = {|t| > tn − 1, a/2} |
|
where

Two samples
Two independent samples t-test with normal populations and σ12 = σ22 unknown
Next table shows the rejection regions of two independent samples t-test with normal populations and σ12 = σ22. ggttest is also used to visualize this test.
| H0 | H1 | Rejection Region of gginference |
Parameters of t.test
|
|---|---|---|---|
| μ1 − μ2 = d0 | μ1 − μ2 < d0 | R = {t < − tn1 + n2 − 2, a} |
|
| μ1 − μ2 > d0 | R = {t > tn1 + n2 − 2, a} |
|
|
| μ1 − μ2 ≠ d0 | R = {|t| > tn1 + n2 − 2, a/2} |
|
where
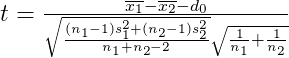
Two independent samples t-test with normal populations and σ12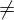 σ22
σ22
ggttest is used to visualize two independent samples t-test with normal populations and σ12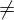 σ22. The following table shows the rejection regions of this test.
σ22. The following table shows the rejection regions of this test.
| H0 | H1 | Rejection Region of gginference |
Parameters of t.test
|
|---|---|---|---|
| μ1 − μ2 = d0 | μ1 − μ2 < d0 | R = {t < − tν, a} |
|
| μ1 − μ2 > d0 | R = {t > tν, a} |
|
|
| μ1 − μ2 ≠ d0 | R = {|t| > tν, a/2} |
|
where
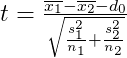
and ν degrees of freedom with

Paired samples with normal population
ggttest is used also to visualize the results of the paired sample Student’s t-test. Next table shows th rejection region of this test.
| H0 | H1 | Rejection Region of gginference |
Parameters of t.test
|
|---|---|---|---|
| μ1 − μ2 = d0 | μ1 − μ2 < d0 | R = {t < − tn − 1, a} |
|
| μ1 − μ2 > d0 | R = {t > tn − 1, a} |
|
|
| μ1 − μ2 ≠ d0 | R = {|t| > tn − 1, a/2} |
|
where

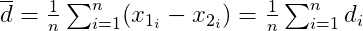

Proportion test
One-proportion z-test
ggproptest() is used to visualize one-proportion z-test. The rejection regions are shown below.
| H0 | H1 | Rejection Region of gginference |
Parameters of prop.test()
|
|---|---|---|---|
| p = p0 | p < p0 | R = {z < − za} |
|
| p > p0 | R = {z > za} |
|
|
| p ≠ p0 | R = {|z| > za/2} |
|
where
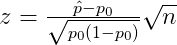
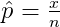
Two-proportion z-test
The results of two-proportion z-test are visualized using ggproptest() and next table shows the rejection regions.
| H0 | H1 | Rejection Region of gginference |
Parameters of prop.test()
|
|---|---|---|---|
| p1 − p2 = d0 | p1 − p2 < d0 | R = {z < − za} |
|
| p1 − p2 > d0 | R = {z > za} |
|
|
| p1 − p2 ≠ d0 | R = {|z| > za/2} |
|
where

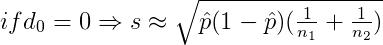


Two-sample F test for equality of variances
ggvartest is used to visualize the results of the paired sample Student’s t-test. The rejection region that is used in this test is shown below.
| H0 | H1 | Rejection Region of gginference |
Parameters of var.test
|
|---|---|---|---|
| σ12 / σ22 = 1 | σ12 / σ22 < 1 | R = {F > Fn1 − 1, n2 − 1, 1 − a} |
|
| σ12 / σ22 > 1 | R = {F > Fn1 − 1, n2 − 1, a} |
|
|
| σ12 / σ22≠ 1 | R = {F > Fn1 − 1, n2 − 1, a/2} |
|
where
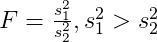
Test for Correlation Between Paired Samples
ggcortest is usesd to visualize the results of test for correlation between paired samples. The following table shows the rejection region of this test.
| H0 | H1 | Rejection Region of gginference |
Parameters of cor.test
|
|---|---|---|---|
| 𝜚 = 0 | 𝜚 ≠ 0 | R = {|t| > tn − 2, a/2} |
|
where
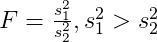
Chi-squared Test of Independence
The results of Pearson’s chi-squared test for count data are visulized using ggchisqtest. Next table shows the rejection region of this test.
| H0 | H1 | Rejection Region of gginference |
Parameters of chisq.test
|
|---|---|---|---|
| Two variables are independent | Two variables are not independent | R = {X2 > χa/22} |
|
where
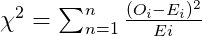
ANOVA F-test
ggaov is used to visualize the results of ANOVA F-test. Table below shows rejection region of Anova F-stest.
| H0 | H1 | Rejection Region of gginference |
Parameters of aov
|
|---|---|---|---|
| H0 : μ1 = μ2= … = μk | Not all three population means are equal | R = {F > Fk − 1, n − k, a} |
|
where
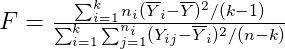
Getting help
If you encounter a bug, please feel free to open an issue with a minimal reproducible example.

